Biodiversity Reading time 6 min
FROGS: a frog attacking fungi
Published on 05 October 2021
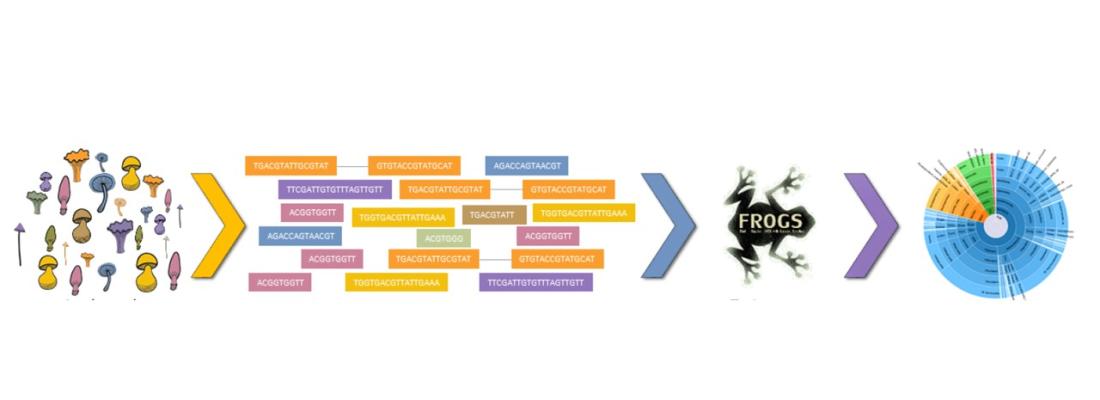
Fungi are present in all environments. They fulfil important ecological functions and play a crucial role in the food industry. Their accurate characterization is thus indispensable, particularly through metabarcoding.
The most frequently used markers to monitor fungi are ITSs. These markers are the best documented in public databases but have one main weakness: polymerase chain reaction amplification may produce non-overlapping reads in a significant fraction of the fungi. When these reads are filtered out, traditional metabarcoding pipelines lose part of the information and consequently produce biased pictures of the composition and structure of the environment under study.
We developed a solution that enables processing of the entire set of reads including both overlapping and non-overlapping, thus providing a more accurate picture of fungal communities. Our comparative tests using simulated and real data demonstrated the effectiveness of our solution, which can be used by both experts and non-specialists on a command line or through the Galaxy-based web interface.
This new version of FROGS is the result of a collaboration between the INRAE laboratories GABI and MaIAGE in Jouy-en-Josas and GenPhySE in Toulouse. It has been described in a scientific article published in Briefings in Bioinformatics and is available on the platforms of http://bioinfo.genotoul.fr/and https://migale.inrae.fr/. FROGS can be used for all types of overlapping amplicons i.e. 16S, 18S, coi, rpob... or non-overlapping i.e. ITS1, ITS2, D1-D2, rpb2...
FROGS codes are accessible and available on https://github.com/geraldinepascal/FROGS and via conda: https://anaconda.org/bioconda/frogs
For more information, please visit the FROGS website: http://frogs.toulouse.inrae.fr
Contact: frogs-support@inrae.fr
Maria Bernard, Olivier Rué, Mahendra Mariadassou, Géraldine Pascal, FROGS: a powerful tool to analyse the diversity of fungi with special management of internal transcribed spacers, Briefings in Bioinformatics, 2021; bbab318
https://doi.org/10.1093/bib/bbab318
