Bioeconomy Reading time 3 min
Understand how bacteria adapt to their environment
Published on 22 March 2021
When a bacterium controls the degradation of its mRNAs for adaptation
A team of researchers from INRAE Occitanie-Toulouse center, in collaboration with Inria Grenoble – Rhône-Alpes, has explored the regulatory mechanisms of mRNA degradation during the adaptation of the Escherichia coli bacterium to its environment. "In both natural and synthetic environments, E. coli cells must cope with changes, such as the availability of carbon sources," explains Muriel Cocaign-Bousquet, INRAE research director. In order to be adapted to the growth environment, bacterial cells use mRNA degradation regulation to favor or, on the contrary, to reduce expression of some genes in order to optimize its cellular function"
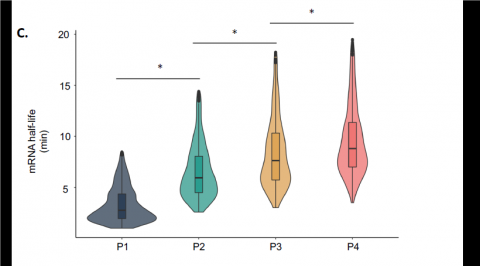
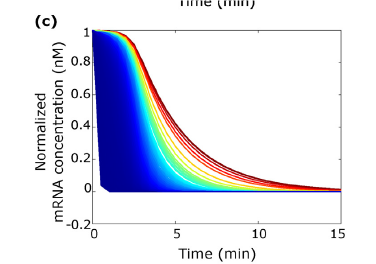
Understanding the adaptation of bacteria to design innovative strains
Using high-throughput biological methods and modeling approaches, researchers at the Toulouse Biotechnology Institute measured the degradation of more than 4000 individual mRNAs in E. coli cells. This allowed to understand how E. coli differently regulates the degradation of certain mRNAs depending on the nature and concentration of the carbon source and to identify 18 major mRNAs for the adaptation of E. coli to its environment. The researchers also characterized the enzymes involved in mRNA degradation and discovered a novel regulatory mechanism related to the competition between thousands of mRNA molecules to bind to an enzyme, the endoribonuclease RNase E.
The better understanding of the adaptation process of E. coli opens up new strategies of mRNA life cycle engineering to improve the performance of this bacterium in biotechnology. By playing with the regulation of mRNA degradation mechanisms and kinetics, it would be now possible to better control gene expression. "This work will be useful to develop innovative strains of E. coli, better adapted to use new carbon sources for the synthesis of proteins and metabolites of interest for the industry", concludes Muriel Cocaign-Bousquet.
Morin M, Enjalbert B, Ropers D, Girbal L, Cocaign-Bousquet M. Genomewide Stabilization of mRNA during a "Feast-to-Famine" Growth Transition in Escherichia coli. mSphere. 2020 May 20;5(3):e00276-20. doi: 10.1128/mSphere.00276-20.
Etienne TA, Cocaign-Bousquet M, Ropers D. Competitive effects in bacterial mRNA decay. J Theor Biol. 2020 Nov 7;504:110333. https://doi.org/10.1016/j.jtbi.2020.110333 Epub 2020 Jun 29. PMID: 32615126.
