Agroecology Reading time 4 min
Dominant and recessive gene expression, chapter Three: back to evolution
Published on 17 December 2014

A linear dominance hierarchy
Researchers first selected six alleles from a Brassicaceae model, Arabidopsis halleri. They then produced several hundred controlled hybrids to obtain an extensive number of allele pairs and determine their phenotypes, enabling the identification of the dominant allele in each pair. These hybrids showed that there is a linear hierarchy in dominance relationships. One allele is dominant over all others, a second is dominant over all alleles except the first, and so on until the last allele, which is recessive in relation to the others.
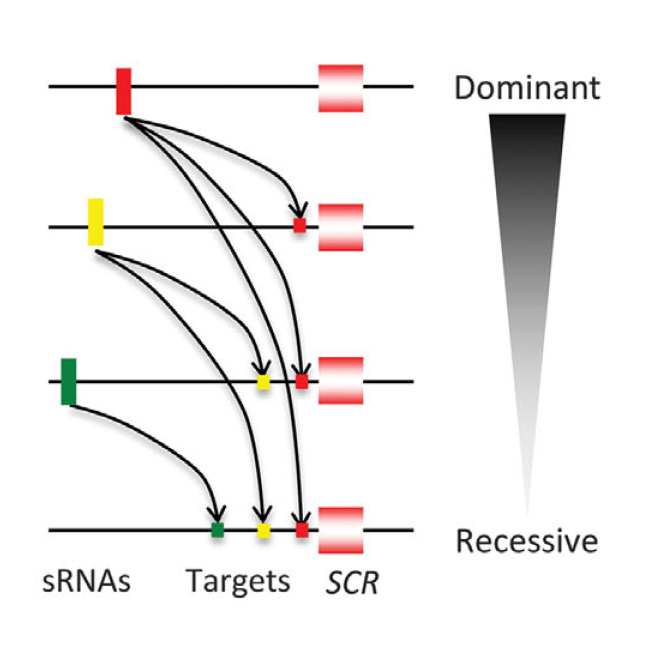
Researchers then used bioinformatics techniques to look for small RNAs and their potential targets on the six selected alleles. They demonstrated that in more than 90% of the dominant relationships observed, there was a small RNA produced by the dominant allele that specifically targeted the recessive allele. The small RNA-target mechanism provides an explanation that is consistent with most phenotypes observed and offers a solid foundation to explain multiallelic dominant relationships.
Putting the theory to the test
The researchers were able to prove their theory by designing an experiment that showed the involvement of the small RNA-target mechanism in genetic dominance. Of the small RNA-target pairs detected in silico, they focused on one in particular and made genetic transformations on Arabidopsis thaliana, a self-pollinating plant (lacking the self-incompatibility mechanism). By importing the self-incompatibility mechanism from Arabidopsis halleri into this plant, researchers were able to show that the presence of small RNA and its target is necessary for a dominant relationship to develop.
An evolutionary mechanism
The researchers extended their analysis to additional alleles. They made in silico predictions about the small RNA sequences and their targets by reconstructing their evolutionary history, and in so doing showed that the duplication, deletion and diversification processes of small RNAs and their targets could explain the development of a dominance network. As hypothesised, it was clear that genetic elements accumulated in different allelic lines. Some alleles have become dominant through the acquisition of small RNAs while others have become recessive through the target acquisition, almost as though they have come to a mutually beneficial agreement. It is as advantageous to be recessive as dominant; what really matters is that heterozygotes express a single allele.
Inter-disciplinary efforts lead to success
Heated debates, mathematical models and theoretic predictions, genetic and phenotypic measurements, functional validations and historical reconstructions: none of these approaches is new in the field of biology. What is exceptional is that all of them were applied simultaneously to study a single biological topic. But working together was necessary to provide a complete explanation. As Dobzhansky famously quipped in 1973, “Nothing makes sense in biology except in the light of evolution.”
