Biodiversity Reading time 2 min
Architect bacteria build networked membranous structures to improve nutrition
PRESS RELEASE - The cereal weevil, a major crop pest, provides a home for symbiotic bacteria within the cells of its body. Scientists from INRAE and INSA Lyon, working with experts from Synchroton SOLEIL and Claude Bernard University in France and with the Max Planck Institute and the European Molecular Biology Laboratory in Germany, have discovered that these bacteria build networks of complex membranous structures inside the host cell, increasing the area of the exchange interface to actively retrieve an essential nutrient – sugar. This is the first time that such enhanced structures created by bacteria have been found. The team’s results, published in Cell, open new research approaches to the understanding of microorganisms, particularly those that inhabit cells, and offer new research opportunities in the fight against crop pests.
Published on 29 October 2025
Cereal weevils are one of the main pests to attack grain crops (wheat, rice, corn) both in the field and during storage. The individual weevil feeds directly on the grain, but within its cells it also carries symbiotic bacteria in large numbers. These bacteria, Sodalis pierantonius, occupy highly specialised cells in their host and generate the essential nutrients that cannot be obtained by the insect from its cereal diet. A mutually beneficial relationship is thus established: the bacteria feed off the carbohydrates (mostly glucose) produced by the insect’s digestion of the cereal and, in exchange, provide it with key vitamins and certain amino acids.
The central importance of these symbiotic nutrient exchanges was already well known to scientists, but the specifics of how such exchanges occur were not. By combining volume electron microscopy with a sample preparation method better able to preserve the membranes, the team was able to observe, for the first time, the presence of abundant and complex tubular membranous networks constructed by the bacteria. To explore the architecture and composition of these newly discovered structures, the scientists then devised observational and analytical methods using 3D microscopy and the Synchrotron SOLEIL particle accelerator facilities1.
‘Tubenets’: the complex exchange networks created by bacteria
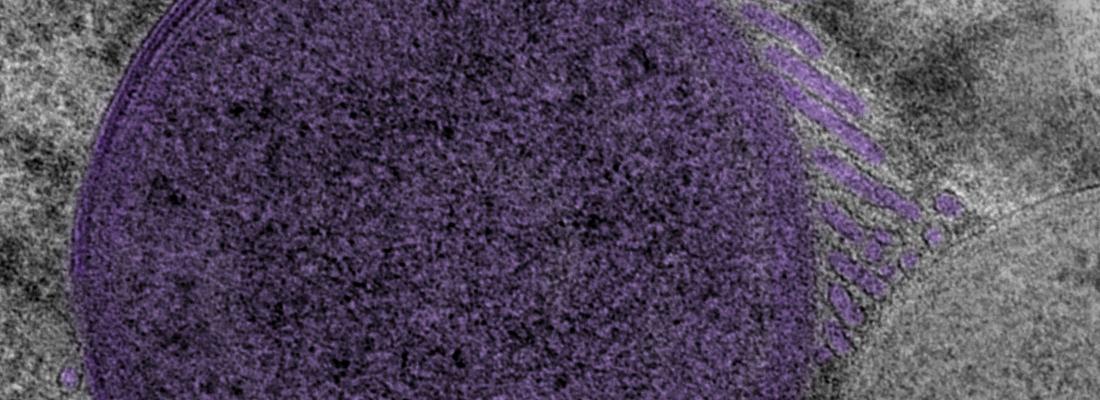
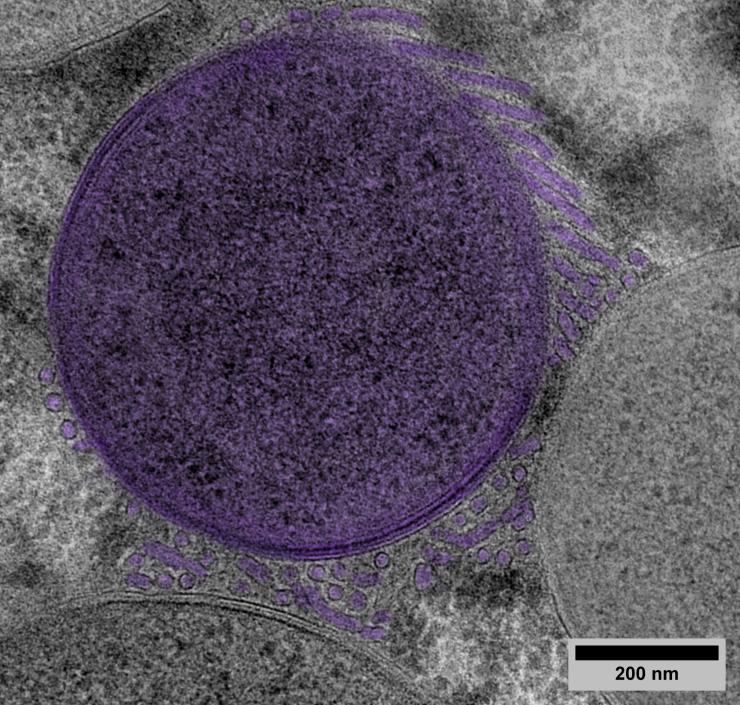
The team’s scientific analysis revealed the presence of structures formed of a complex network of tubes of 0.02 µm in diameter and several µm in length. These created tri-dimensional nets of multiple connections, not only between the bacteria themselves but also with the host. Just as the structure of microvilli2 in the human gut increases nutrient absorption during digestion by expanding the available surface area, these tubular structures enable the bacteria to increase the area of interaction with the host cell to improve their carbohydrate acquisition. In exchange, the bacteria produce key nutrients for the benefit of the insect. The research team have named these structures ‘tubenets’ to reflect their architecture.
While the structures that increase the exchange interface for nutrient absorption in multicell organisms (such as the intestine or plant roots) are familiar to scientists, this is the first time that such a structure has been revealed to exist in bacteria. It is possible that similar structures are to be found in other types of bacteria.
[1] An ultra-high-tech facility, SOLEIL produces synchrotron radiation, an extremely bright light that can be used to explore inert or biological matter down to the atomic scale, thanks to the extremely intense beams of the Synchrotron SOLEIL particle accelerators. In particular, the team used STXM technology, which allows the carbon content of a sample to be determined at very high spatial resolution (30 nm).
[2] Microscopic bristle-like protrusions on the surface of some organs such as the bile duct and the small intestine, through which external materials such as nutrients are absorbed.
Reference
Balmand S. et al. (2025) Bacterial tubular networks channel carbohydrates 1 in insect endosymbiosis. Cell DOI : https://doi.org/10.1016/j.cell.2025.10.001
