Food, Global Health Reading time 3 min
A novel antibiotic resistance mechanism
Published on 21 December 2018
In their natural environment, bacteria coexist with a wide range of microorganisms. To survive and access food, these bacteria have no other choice but to repel or even eliminate their competitors by producing antibiotics. So bacteria also need to be able to protect themselves. They have therefore developed a wide variety of defense mechanisms to combat antibiotics: some have a membrane that is "impermeable" to antibiotics, others pump the antibiotics out, and still others modify them or cut them to inactivate them."We have built up a relatively clear picture of the resistance mechanisms employed by bacteria, but, as our results show, there are probably many mechanisms that we don't even know exist," explains Mélodie Duval, a postdoctoral fellow in the Bacteria-Cell Interactions Unit at the Institut Pasteur. "Two years ago, in collaboration with Rotem Sorek's Israeli team at the Weizmann Institute of Science, we identified a first resistance gene in Listeria that was induced in response to antibiotics,"1continues Pascale Cossart, head of the unit and coordinator of the IBEID LabEx. "The regulation of its expression was unusual but its mechanism of action was fairly typical. This time, the gene that we have recently discovered and characterized induces a totally new resistance mechanism, which I find fascinating," says the scientist.
Once again, it all started with the analysis method developed by Rotem Sorek's team2. This technique, known as "term-seq", reveals the length and abundance of messenger RNA (mRNA) in a given sample, and therefore the degree of expression of the corresponding genes, in different experimental conditions. "We tested the effect of two antibiotics – lincomycin and erythromycin – on Listeria monocytogenes bacteria," explains Mélodie Duval. The bacteria were cultured, with and without antibiotics, then their mRNA was extracted. The findings showed that some mRNA with premature transcription termination in the absence of antibiotics underwent complete transcription in the presence of antibiotics, thereby revealing the induction of some genes."One of these genes especially caught our attention because it was highly analogous to a gene known to help E. colibacteria resist heat shock by splitting their stalled ribosomes in two," says Mélodie Duval. "This particularly interested us because both the antibiotics used in our study, like heat shock, are known to stall bacterial ribosomes." Was there a similarity in the mechanism of action?
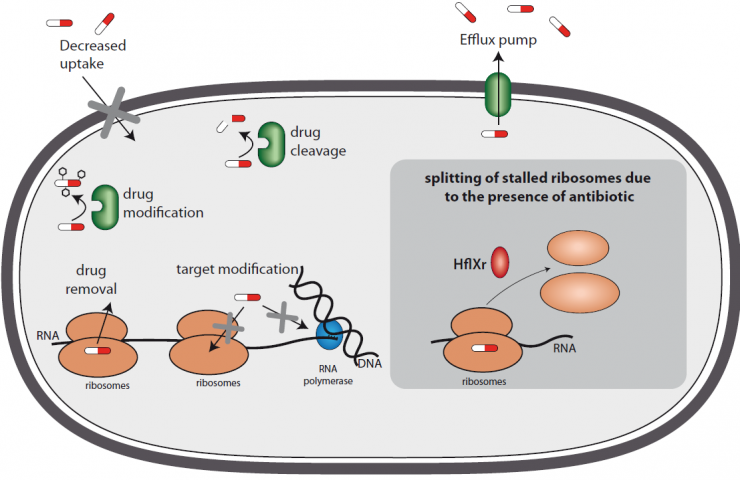
When the scientists examined this gene in more depth, they noticed that the protein it encoded acted on the ribosomes – whose task is to translate mRNA into proteins – and they named ithflXr. Its expression, regulated by an attenuation mechanism3and induced by antibiotics, produces a protein that literally splits the ribosomes in two. But far from destroying them, this actually enables the two ribosome subunits to be recycled and to resume their task of protein synthesis, a process that is vital for bacterial growth. "This mechanism of ribosome splitting to resist an antibiotic is a completely new discovery," confirms Pascale Cossart. And it is likely that the resistance mechanism is not unique to Listeria, since the hflXr gene, and potentially the associated mechanism, is found in a huge number of bacteria, especially in Firmicutes. "As we were carrying out our research, Croatian and Canadian scientists who were searching for antibiotic resistance genes in soil samples taken near an antibiotic manufacturing site and a livestock farm revealed that they had found the hflX gene4. It is satisfying to know that other scientists have detected in the environment what we have discovered and elucidated in Listeria in the laboratory. It neatly confirms the results of our basic research," concludes Mélodie Duval.
This research received funding from the Institut Pasteur, Inserm, INRA, the ERC (BacCellEpi Advanced Grant), the Weizmann-Pasteur Institute, the Le Roch-Les Mousquetaires Foundation and the IBEID LabEx.
1 Dar D, Shamir M, Mellin JR, et al (2016) Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:1–12 . DOI : 10.1126/science.aad9822
2 Duval M, Dar D, Carvalho F, Rocha E.P.C., Sorek R and Cossart P, HflXr, an homolog of a ribosome-splitting factor, mediates antibiotic resistance.Proc. Natl. Acad. Sci USA 2018, in press
3 A mechanism for regulating gene expression present in bacteria, where transcription terminates prematurely, upstream of the structural gene, and therefore no expression occurs.
4 González-Plaza JJ, Šimatovic A, Milakovic M, et al (2018) Functional Repertoire of Antibiotic Resistance Genes in Antibiotic Manufacturing Effluents and Receiving Freshwater Sediments. Front Microbiol 8:1–13 . DOI : 10.3389/fmicb.2017.02675
5 Lau CH-F, Van Engelen K, Gordon S, et al (2017) Novel antibiotic resistant gene from agricultural soil exposed to antibiotics widely used in human medicine and animal farming. Appl Environ Microbiol 83:1–18 . DOI : 10.1128/AEM.00989-17
Mélodie Duval, Daniel Dar, Filipe Carvalho, Eduardo P. C. Rocha, Rotem Sorek, and Pascale Cossart HflXr, a homolog of a ribosome-splitting factor, mediates antibiotic resistance, PNAS, 13 decembre 2018
