Reading time 6 min
Looking for a needle in a haystack: de novo phenotypic target identification reveals Hippo pathway-mediated miR-202 regulation of egg production
Published on 08 December 2023
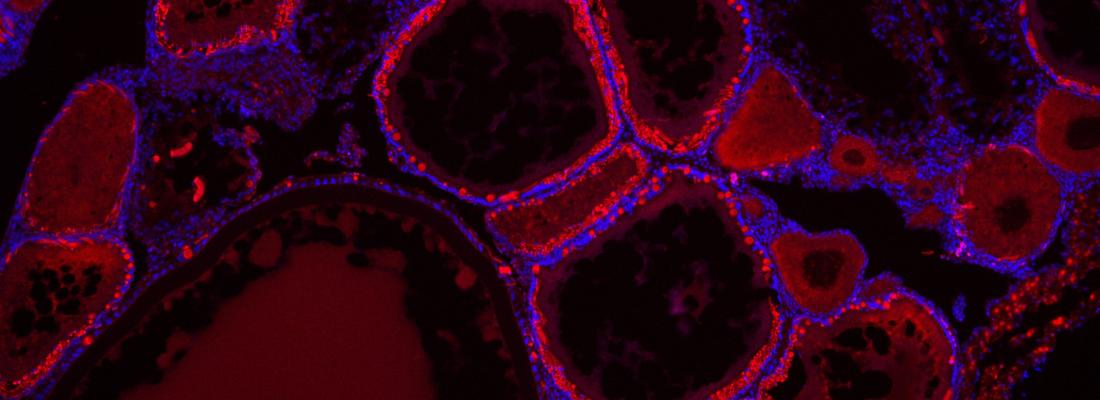
Understanding microRNA (miRNA) functions has been hampered by major difficulties in identifying their biological target(s). Currently, the main limitation is the lack of a suitable strategy to identify biologically relevant targets among a high number of putative targets. Here we provide a proof of concept of successful de novo (i.e. without prior knowledge of its identity) miRNA phenotypic target (i.e. target whose de-repression contributes to the phenotypic outcomes) identification from RNA-seq data.
Using the medaka mir-202 knock-out (KO) model in which inactivation leads to a major organism-level reproductive phenotype, including reduced egg production, we introduced novel criteria including limited fold-change in KO and low interindividual variability in gene expression to reduce the list of 2853 putative targets to a short list of 5. We selected tead3b, a member of the evolutionarily-conserved Hippo pathway, known to regulate ovarian functions, due to its remarkably strong and evolutionarily conserved binding affinity for miR-202-5p. Deleting the miR-202-5p binding site in the 3′ UTR of tead3b, but not of other Hippo pathway members sav1 and vgll4b, triggered a reduced egg production phenotype.
This is one of the few successful examples of de novo functional assignment of a miRNA phenotypic target in vivo in vertebrates.
This work is the result of collaboration between Julien Bobe's team at the INRAE Fish Physiology and Genomics Institute in Rennes and Hervé Seitz's team at the CNRS in Montpellier, and is part of the ANR-funded MicroHippo project coordinated by Julien Bobe.
Janati-Idrissi S, de Abreu MR, Guyomar C, de Mello F, Nguyen T, Mechkouri N, Gay S, Montfort J, Gonzalez AA, Abbasi M, Bugeon J, Thermes V, Seitz H, Bobe J. Looking for a needle in a haystack: de novo phenotypic target identification reveals Hippo pathway-mediated miR-202 regulation of egg production. Nucleic Acids Res. 2023 Dec 7:gkad1154. http://dx.doi.org/10.1093/nar/gkad1154
