Agroecology Reading time 3 min
The first sunflower resistance gene identified: towards a more efficient control of the parasitic plant Orobanche cumana
Published on 17 September 2019
Sunflower is a crop with many advantages for both human food and non-food uses and and for its agronomic and environmental characteristics. It is a major Crop for the French oil and protein industry.
One of the main constrain for sunflower are due to pests that impact yield, such as phoma, downy mildew or the parasitic plant Orobanche cumana.
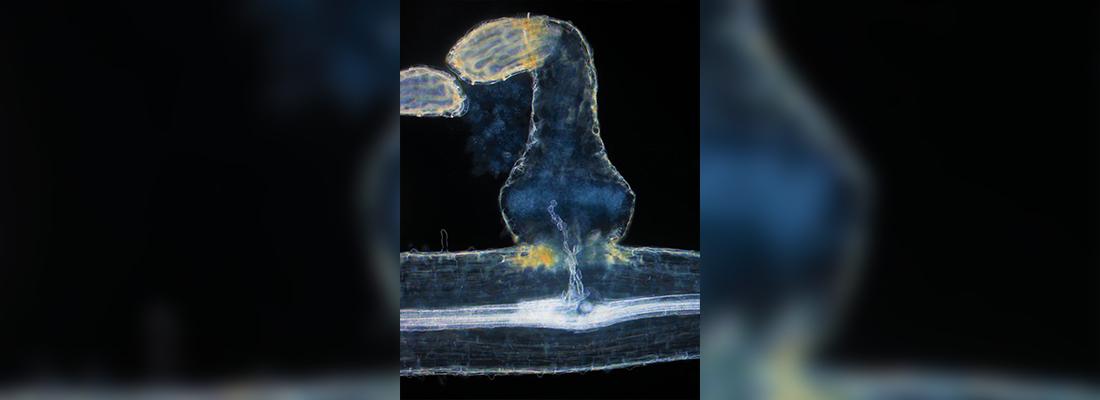
Parasitic plants, such as Orobanche cumana, depend on other plants to complete their life cycle. They attach either to the stems or the roots of the host, connecting through an organ that enable to penetrate in the tissues and then absorb water and nutrients, becoming a new sink for the host.
Sunflower broomrape is an obligate parasitic plant that connects the roots of the sunflower, causing strong yield losses in extreme cases.
Understanding the molecular mechanisms of plant-plant interactions
Plants interact with each other, but the mechanisms of the interaction between plants are still poorly understood.
In order to understand the resistance mechanisms of sunflowers, it is needed to look at the sunflower DNA and locate the trait/gene that confers these resistances.
To map the HaOr7 gene on the sunflower genome, different genetic mapping strategies were used using susceptible and resistant sunflowers containing the HaOr7 gene. The gene was precisely located on chromosome 7.
The analysis showed that HAOR7 is a membrane receptor that transduce a signal when O. cumana attaches to the roots and prevents the connection of the parasitic plant. In susceptible sunflowers, this protein is not functional, which allows the parasitic plant to connect to the sunflower root and to develop.
At what stage of the interaction does the gene act?
The life cycle of O. cumana has four main stages, the first three are underground: (1) induction of seed germination, (2) penetration of the parasite into the roots of the host, (3) formation of a tubercle (4) emergence of a single stem from this tubercle from the soil, producing several flowers that can each produce hundreds of seeds. Up to a hundred broomrape stems can grow on a single sunflower.
The authors were able to show that susceptible and resistant sunflowers were able to induce the germination of the O. cumana seeds similarly, but in resistant sunflowers, O. cumana was unable to connect to the vascular system of the sunflower roots. Thus, without water and nutrients, they die rapidly, without developing nor tubercles and nor floral stems.
These results will improve the resistance of the varieties to sunflower broomrape and thus enable farmers to maintain yields.
This work was carried out with the support of the National Center for Plant Genomic Resources, (CNRGV) in collaboration with Syngenta, and the participation of El Instituto de Agricultura Sostenible (IAS) del Consejo Superior de Investigaciones Científicas (CSIC) in Cordoba, Spain.
Full reference of the publication : A receptor-like kinase enhances sunflower resistance to Orobanche cumanaPauline Duriez, Sonia Vautrin, Marie-Christine Auriac, Julia Bazerque, Marie-Claude Boniface, Caroline Callot, Sébastien Carrère, Stéphane Cauet, Mireille Chabaud, Fabienne Gentou, Marta Lopez-Sendon, Clémence Paris, Prune Pegot-Espagnet, Jean-Christophe Rousseaux, Begoña Pérez-Vich, Leonardo Velasco, Hélène Bergès, Joël Piquemal & Stéphane Muños, Nature Plants, volume 5, pages 1211–1215 (2019) doi:10.1038/s41477-019-0556-z. |

