Ressources dossier
Food, Global HealthMetaGenoPolis, a unique place dedicated to knowledge on microbiota
Published on 05 October 2022

Inside the labs : 5 stages, from sample collection to knowledge production
> Sample collection and management
Whether in the context of clinical trials or research projects, the first step is the collection of stool samples from cohorts or individual volunteers. For this purpose, and as part of the International Human Microbiome Standards (IHMS) project that it coordinates, MetaGenoPolis has developed standard procedures that are a global reference for the preparation of stool samples, aimed at guaranteeing their integrity and that of their extracted DNA for a robust analysis of the microbiota.
> The biobank, management and storage facility for faecal samples
Once the samples have been obtained, aliquots are generated, each with a barcode to identify them. These aliquots are then stored securely in the biobank. Fully automated, it can store up to 600,000 aliquots and DNA extracts at -80°C. A robotic arm deposits or retrieves the tubes for each analysis or experiment.
> Preparation of DNA extracted from samples for sequencing
Before the DNA of the microorganisms present in the samples can be sequenced and thus characterised, it is necessary to extract this DNA by separating it from other cellular components and residues present in the samples. This involves physical and chemical disruption steps as well as proven and standardised separation techniques.
> High-throughput sequencing of DNA from microorganisms
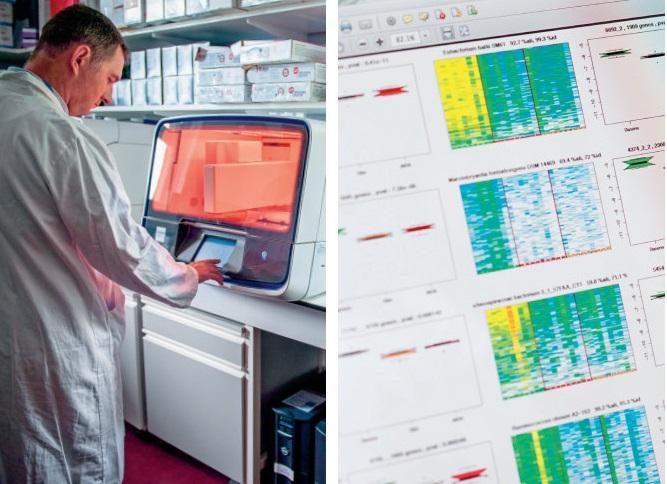
High-throughput sequencing of the DNA of microorganisms (mainly bacteria) living in the digestive tract allows the characterisation of the gut microbiota. MetaGenoPolis uses the whole genome sequencing method called “shotgun metagenomics”, which focuses on all the genes of the accessible microorganisms. Using a reference gene database, the DNA fragments read by the sequencer identify the genes of the microorganisms present in the aliquot and their proportion. A considerable volume of computer data is thus generated and analysed.
> Computational and biostatistical analysis
After sequencing the DNA, scientists obtain a large amount of data which is then analysed to answer different questions: for example, to study the impact of the consumption of a product, of a diet or of a probiotic on the gut microbiota. Cross-referencing data from individuals in the same cohort can help identify the role of the microbiota in diseases such as obesity, diabetes, inflammatory diseases or liver cirrhosis.
> Screening platform for functional metagenomic analysis
This automated high-throughput screening platform makes it possible to analyse interactions between microorganisms and epithelial or immune cell lines. Functional metagenomics thus allows a better understanding of the function of each of the bacteria in the gut microbiota, to decode host-microbiota interactions and to identify new target molecules of therapeutic interest.
MetaGenoPolis in figures
140
scientific publications, many of them in high impact journals.
15 of these publications are among the most cited in the world.
36
patents.
10
licences.
82
research projects with more than 100 public partners (including 8 European projects and 5 international projects) for a total budget of 18 million euros.
154
research projects for innovation with 69 private partners in the agrifood and health sectors, for a total budget of
24 million euros.
-
Elodie Regnier
Author / Translated by Alessandra Riva
